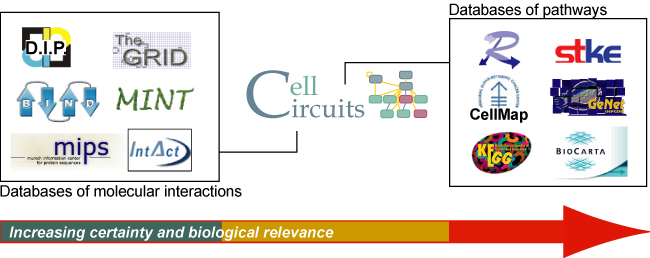
An open-access database of molecular network models that:
- Bridges the gap between databases of individual pair-wise
molecular interactions and databases of validated pathways.
- Contains functional network hypothesis produced by algorithms that screen molecular interaction networks based on their correspondence with expression or phenotypic data, their internal structure, or their conservation across species.
- Is searchable using protein/gene names and Gene Ontology terms. Models are available either as images or in machine-readable formats.
- Serves as a clearinghouse in which theorists may distribute or revise models in need of validation and experimentalists may search for models or specific hypotheses relevant to their interests.
|
|
Search the network models in CellCircuits ...
(try entering the words in underline bold into the search box or click on the examples below)
- By gene
- Using gene names or aliases (registered by the GO database) (e.g. GCN4)
- Models in CellCircuits contain genes from yeast (S. cerevisiae), fly (D. melanogaster), worm (C. elegans), human (H. sapiens), and the malaria parasite (P. falciparum)
- An asterisk (*) is a wild card (e.g. GCN*)
- Enter more than one gene, each separated by a space. (e.g. YAP1 SOD1)
- By Gene Ontology annotation
- Use GO ID numbers (e.g. GO:0003677)
- Use partial or complete GO term names (enclosed in double quotes) (e.g. "DNA binding")
|
|
CellCircuits Home |
Advanced Search |
Help |
Ideker Lab |
UCSD
Funding provided by the National Science Foundation (NSF 0425926).
|